Optimize Your Therapeutic Proteins Digitally
We design optimized protein and peptide variants using proprietary AI. You only test the best 30 to 96 candidates in your lab. Faster discovery, fewer experiments.

We design optimized protein and peptide variants using proprietary AI. You only test the best 30 to 96 candidates in your lab. Faster discovery, fewer experiments.
A simple, efficient lab-in-the-loop process. You provide the protein, we deliver optimized variant designs ready for lab validation.
Provide us with your parental protein or peptide sequence and define the optimization goals: affinity, stability, immunogenicity, or multiple properties at once.
Our proprietary algorithm generates millions of candidates and selects the top 30-96 variants optimized for your target properties. Works from just a sequence alone.
You test the designed variants in your lab. If the goal isn't reached, we run additional optimization rounds incorporating your experimental data for even better results.
Our proprietary algorithm combines protein large language models with evolutionary optimization to navigate the vast sequence space efficiently.
Works on any protein or peptide: antibodies, nanobodies, DARPins, peptides, and more. No structural constraints on input.
Optimize any measurable property: binding affinity, aggregation, thermal stability, immunogenicity, selectivity, or multiple at once.
Combine with NGS from ribosome display, site saturation mutagenesis results, docking experiments, or your own property prediction models.
All we need is the parental amino acid sequence. Additional data helps but is not required to start optimizing.
Size Exclusion Chromatography comparison of a parental protein and the best nuantic-optimized variant, from a project with Molecular Partners.
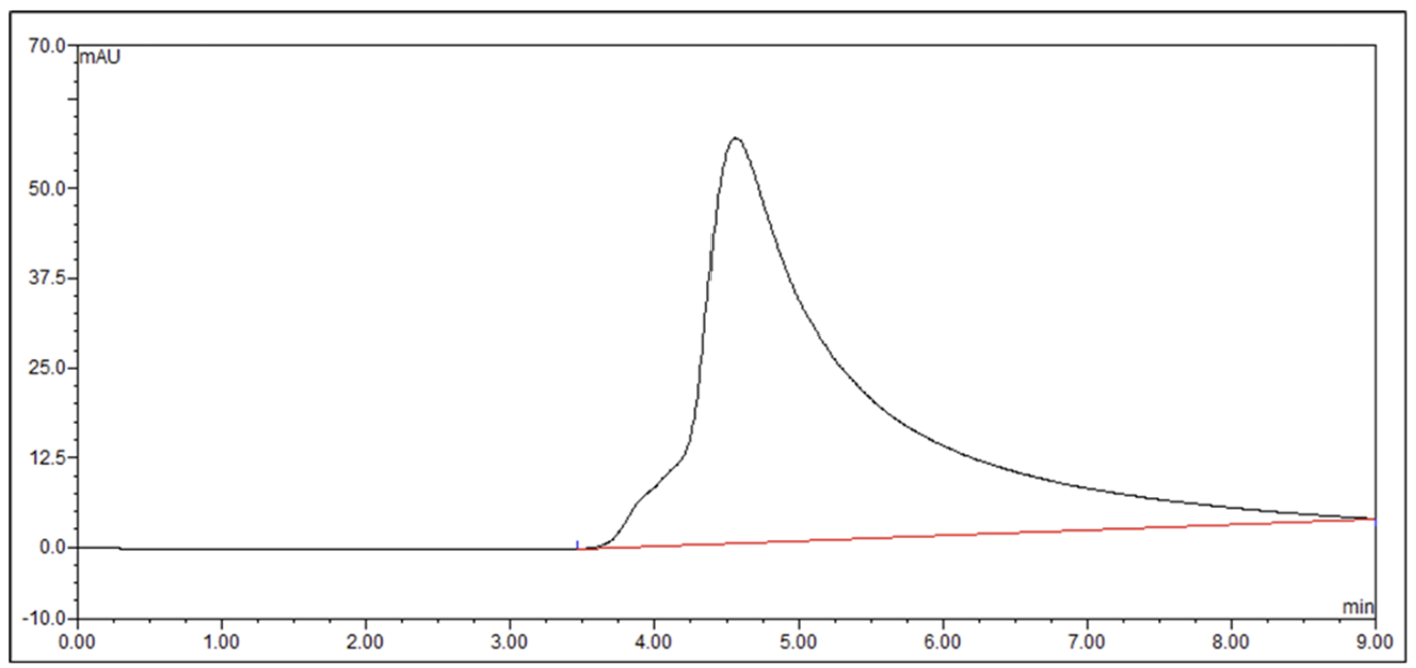
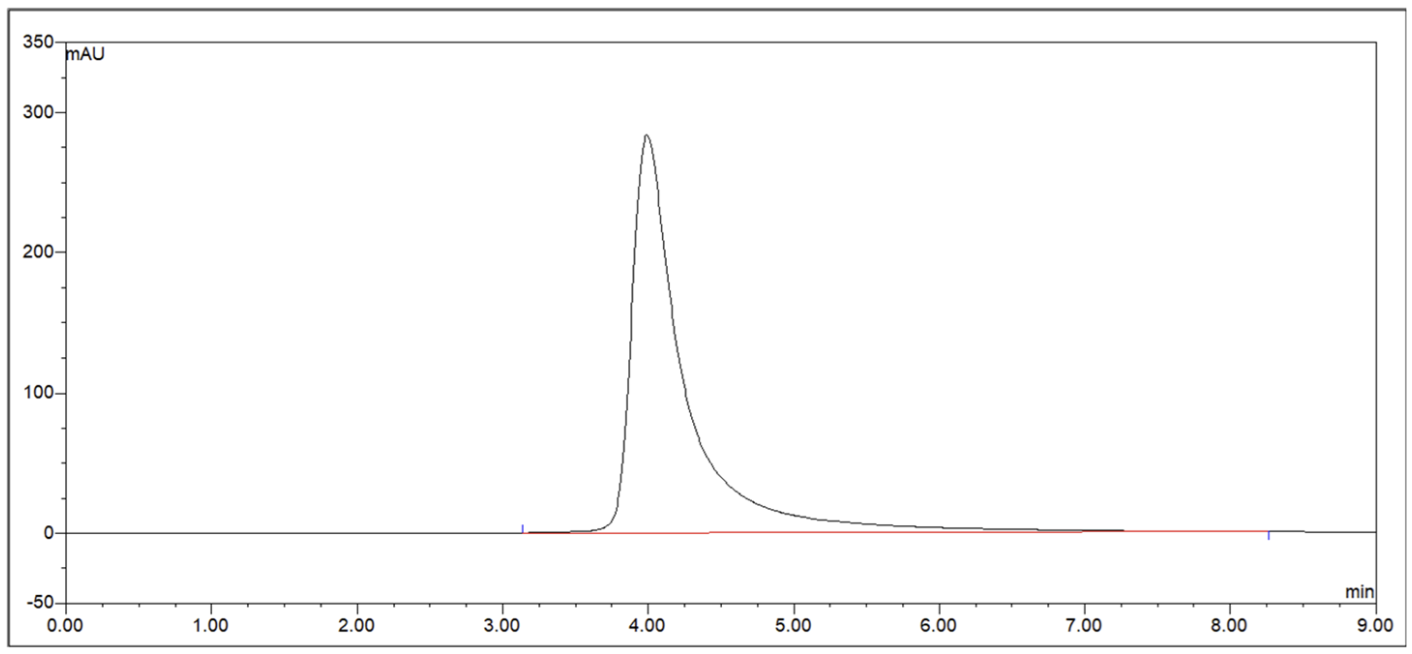
Aggregation completely removed with only 30 designed variants, while maintaining double-digit picomolar binding (~30 pM). Data shown with permission from Molecular Partners (blinded).
Demonstrated across five pilot projects, covering a range of optimization objectives.
400 nM to 150 pM when combining our algorithm with single point mutations from Site Saturation Mutagenesis. Our 30 designs outperformed 48 rational design variants.
400 nM to 14 nM in one optimization round with just 92 designed variants, no additional experimental input needed.
Eliminated predicted T-cell epitopes (NetMHCPan 4.1) while fully maintaining function and favorable biophysical properties.
Tackled multiple biophysical issues in parallel, producing an improved parental used on 2 out of 3 candidates in a lead panel for drug discovery.
25+ combined years at Molecular Partners in protein engineering and optimization. We've lived the problems we solve.






VentureKick Stage 2 completed • Swiss Innovation Challenge 7th out of 100+ startups • Bluelion Alumni • Microsoft for Startups program member • 5 pilot projects with Molecular Partners
Tell us about your project and optimization goals. We'll get back to you to discuss how our technology can accelerate your drug discovery pipeline.
Send Us an Email →Your sequence and project details are treated as strictly confidential.